Predicting the structure and dynamics of membrane protein GerAB from Bacillus subtilis
12 April 2021
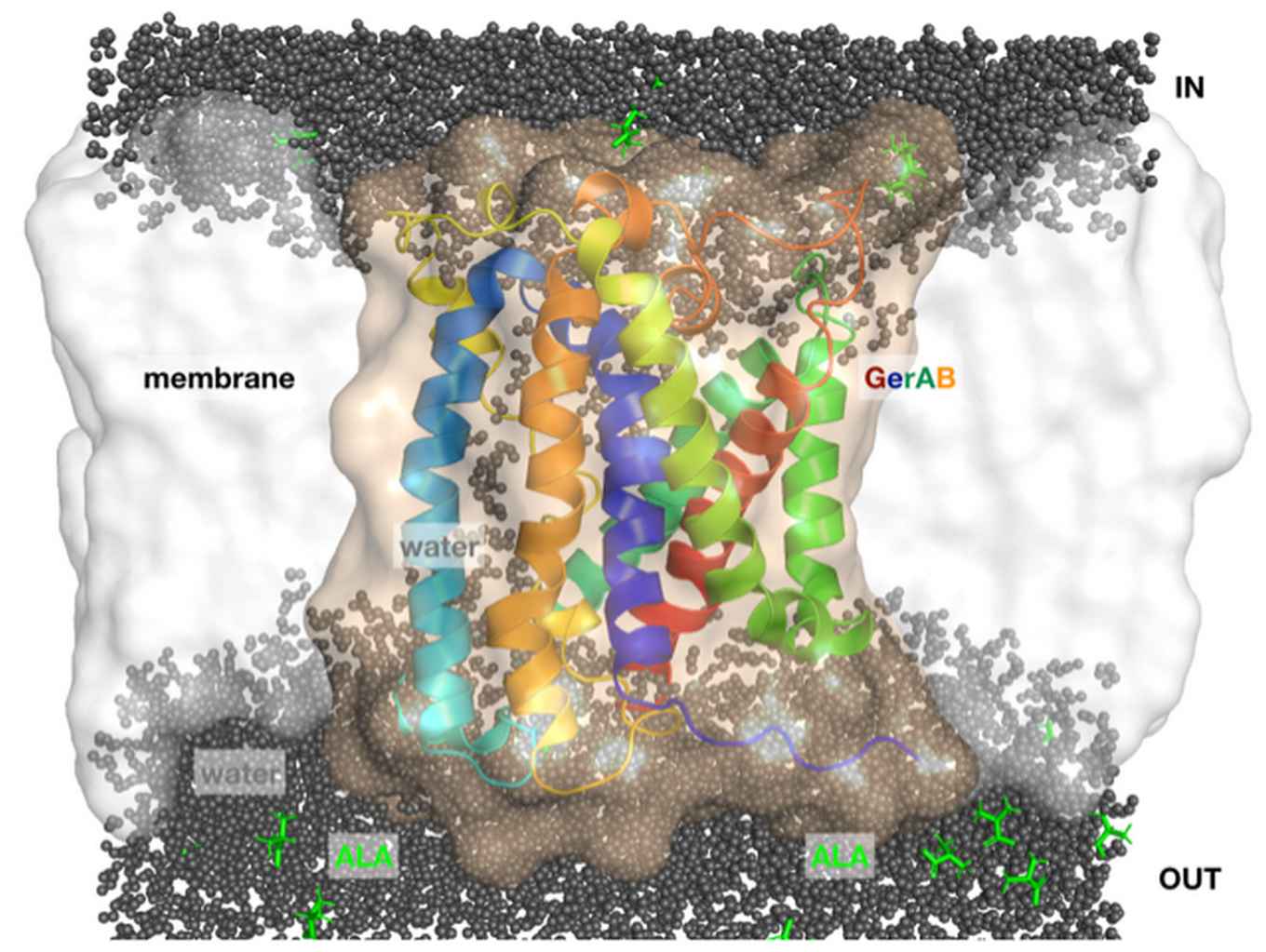
A paper describing the study has just been published in the International Journal of Molecular Sciences. First author is Sophie Blinker, a talented student doing a double masters in both Biomedical Sciences and Medicine at the UvA. She started the study on the germination mechanisms of Bacillus subtilis as part of her BSc research honours excellence track and pursued this during student internships under the supervision of Prof. Stanley Brul of SILS.
Abstract
Bacillus subtilis forms dormant spores upon nutrient depletion. Germinant receptors (GRs) in the spore’s inner membrane respond to ligands such as L-alanine, and trigger spore germination. In B. subtilis spores, GerA is the major GR, and has three subunits, GerAA, GerAB, and GerAC. L-Alanine activation of GerA requires all three subunits, but which subunit binds L-alanine is unknown. To date, how GRs trigger germination is unknown, in particular due to a lack of detailed structural information about the GerAB subunits. Using homology modelling with molecular dynamics (MD) simulations, we present structural predictions for the integral membrane protein GerAB. These predictions indicate that GerAB is an a-helical transmembrane protein containing a water channel. The MD simulations with free L-alanine show that alanine binds transiently to specific sites on GerAB. These results provide a starting point for unravelling the mechanism of L-alanine mediated signalling by GerAB, which is crucially involved in the early events of bacterial spore germination.
Publication details
Blinker, S.; Vreede, J.; Setlow, P.; Brul, S. Predicting the Structure and Dynamics of Membrane Protein GerAB from Bacillus subtilis. Int. J. Mol. Sci. 2021, 22, 3793. DOI: 10.3390/ijms22073793
Links
HIMS research group Computational Chemistry
SILS research group Molecular Biology and Microbial Food Safety